Model Discovery Analysis
Universal Differential Equations (UDEs)[1] have emerged as a promising approach for integrating experimental data into mechanistic models via neural networks.
In this analysis, we make the neural network available as a component that can be plugged in to your existing model. You can then train that neural network using the NNTrainingAnalysis, and then estimate the possible equations that the neural network encodes using the SymbolicRegressionUDEAnalysis.
While the original UDE methodology was developed for systems of ordinary differential equations, acausal modeling presents a different challenge: we begin with differential-algebraic equations (DAEs) for each component, which are then condensed through structural simplification into a final DAE system for solution.
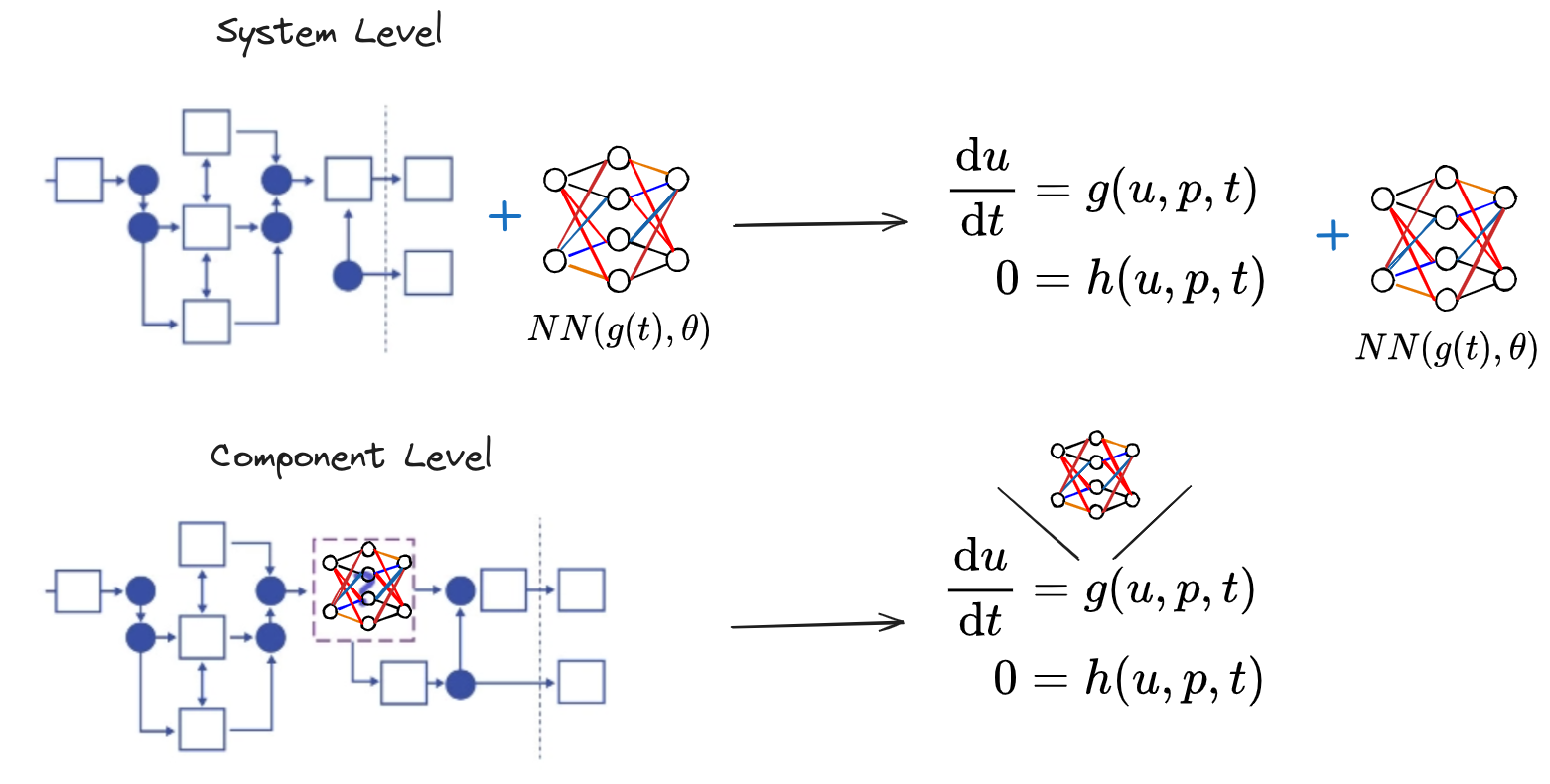
When formulating UDEs in the DAE context, we can operate at two different stages: before structural simplification (component UDE formulation) or after simplification on the resulting DAE system (system UDE formulation).
The component UDE formulation offers significant advantages. By representing the model discovery process at the block component level, we can target specific, deeply nested parts of a model while maximizing reuse of existing structural information. This approach ensures that discovered models remain expressible in the same format as the original model.
Dyad's component UDE analysis enables symbolic representation of neural networks at the block diagram level, allowing us to leverage existing system structure knowledge throughout the model discovery process.
Method Overview
We will embed a (single) neural network inside the system (represented as NeuralNetworkBlock), such that the predictions or the outputs of the neural network affect the dynamics in the desired way based on its inputs.
The Dyad compiler uses the physical description of the system, which maps to the block diagram representation, to generate a system of differential-algebraic equations via structural simplification:
For the component UDE model discovery analysis, we would like to perform a transformation on the system before structural simplification. In this case the system is represented by several systems of DAEs that are connected to each other. These systems can be also represented graphically via block diagrams, where each block is a potentially nested system of DAEs. When we perform the system transformation, we will embed a (single) neural network inside the system (represented as NeuralNetworkBlock), such that the predictions or the outputs of the neural network affect the dynamics in the desired way based on its inputs.
The NeuralNetworkBlock is represented as the following nonlinear system
where inputs and outputs are vector variables that can be used throughout the system as desired and nn_p are the parameters of the neural network.
After we assemble the system and structurally simplify it, we will obtain a new system,
If we use
Example Workflow
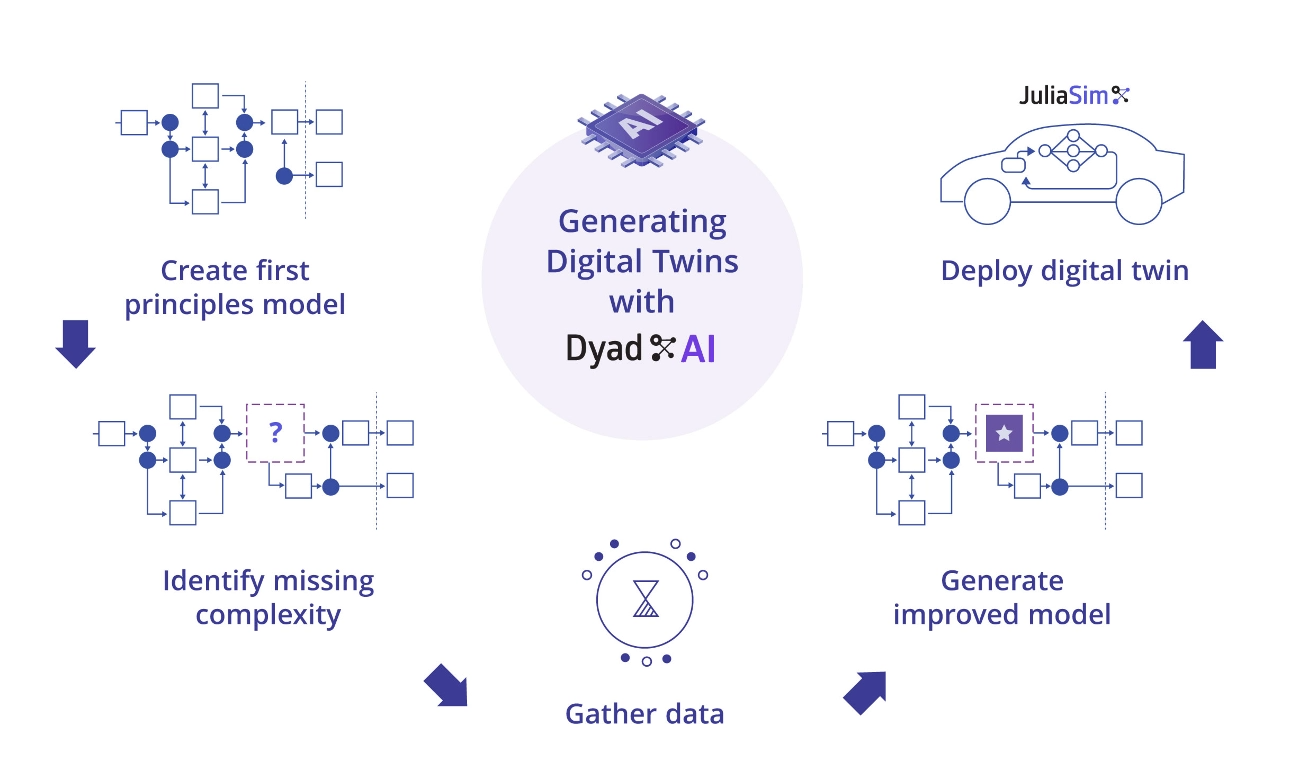
To run a model discovery analysis, we need a model and some data that corresponds to the behaviour that we would want from it. In this example we will consider a simple thermal system where we know part of the structure, but not enough to account for the behaviour of the system. We will add a neural network to the system, train it and then perform symbolic regression to recover possible candidate terms for the missing physics.
using Main.var"##build/.dyad/analyses/udesDyadHygiene#301".component_ude # hide
using DyadInterface, DyadData, Plots # hide
sol0 = SimplePotTransient() # hide
model = artifacts(sol0, :SimplifiedSystem) # hide
data = DyadTimeseries("dyad+juliahub://juliahub.com/datasets/juliasimtutorials/potplate", independent_var = "timestamp", dependent_vars = ["pot.T(t)"]) # hide
tbl = build_table(data) # hide
plt = plot(sol0, idxs=model.pot.T, label = "Initial system") # hide
scatter!(tbl.timestamp, tbl.var"pot.T(t)"; label = "Real data") # hide
plt # hideThis example requires the following julia packages to be installed in the current environment. Use the Julia REPL to add them:
using Pkg
Pkg.add(["ThermalComponents", "BlockComponents", "DyadData", "DyadModelDiscovery"])Let's first create the starting model in Dyad. If you already have a component library, you can add this into a new .dyad file in the dyad folder, otherwise you need to create a new component library via the Command Palette in VS Code (⇧ ⌘ P on MacOS, Ctrl-Shift-P on Windows). See the getting started instructions for more details.
Dyad code
A common issue in modeling is failing to account for all the possible ways in which a system could be storing energy, or otherwise have memory. Such behaviour is encoded by additional state variables that might be missing in a model, and thus it would not match up with the observed behavior. In this example we will study the case of a thermal capacitance for a plate, but other scenarios include elastic energy stored in parasitic flexibilities in mechanical systems, parasitic capacitance and inductance in electrical circuits, time delays in digital communication etc.
In the following we consider a pot that is being heated and some real world measurements of that system. But the dynamics of the initial model does not match the data! Let's use UDEs to discover those dynamics.
We can now add the following Dyad code describing a pot (modelled as a HeatCapacitor) that is being heated:
component SimplePot
parameter C2::HeatCapacity = 15
heat_input = BlockComponents.Sine(amplitude=1, frequency=1/(2*pi))
source = ThermalComponents.PrescribedHeatFlow(T_ref=273.15)
pot = ThermalComponents.HeatCapacitor(C=C2, T0=273.15)
air = ThermalComponents.ThermalConductor(G=0.1)
env = ThermalComponents.FixedTemperature(T=293.15)
Tsensor = ThermalComponents.TemperatureSensor()
relations
connect(heat_input.y, source.Q)
connect(source.node, pot.node)
connect(pot.node, Tsensor.node)
connect(pot.node, air.node_a)
connect(air.node_b, env.node)
end
analysis SimplePotTransient
extends TransientAnalysis(stop=100.0)
model = SimplePot()
end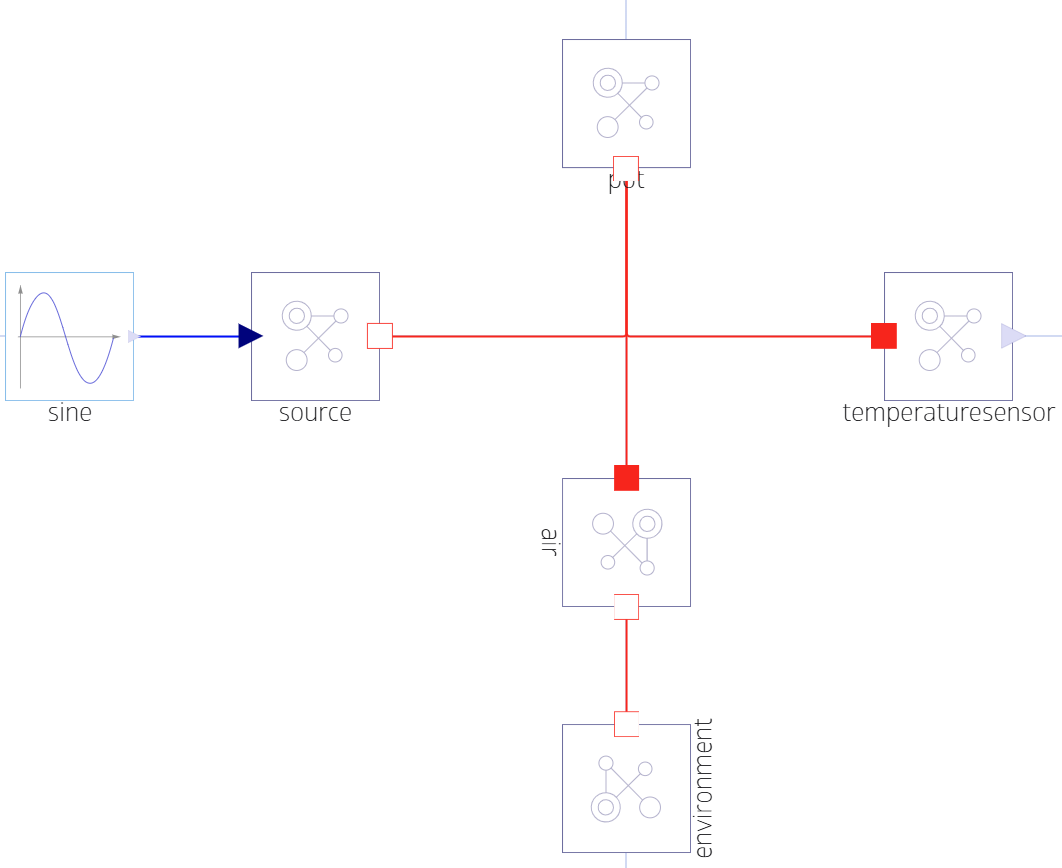
Let's start by running a transient analysis for the above model and check how it compares against the desired system behaviour.
using Main.var"##build/.dyad/analyses/udesDyadHygiene#301".component_ude # hide
using DyadInterface, DyadData, Plots
sol0 = SimplePotTransient()
model = artifacts(sol0, :SimplifiedSystem)
data = DyadTimeseries("dyad+juliahub://juliahub.com/datasets/juliasimtutorials/potplate", independent_var = "timestamp", dependent_vars = ["pot.T(t)"])
tbl = build_table(data)
plt = plot(sol0, idxs=model.pot.T)
scatter!(plt, tbl.timestamp, tbl.var"pot.T(t)"; label = "Real data")
pltWith the component UDE approach, we want to add a new component in the model that would be connected between the source and the pot such that we can use it to discover the missing physics. To this end, we will build a ThermalNN component that adds a new state in the system (x) that is prescribed by the output of the neural network, but it also incorporates physics knowledge about the system by formulating the component as something that outputs a heat flow rate based on some scaled input temperatures.
component ThermalNN
structural parameter n_input::Integer = 2
structural parameter n_output::Integer = 1
nn = DyadModelDiscovery.NeuralNetworkBlock(n_input=n_input, n_output=n_output, chain=DyadModelDiscovery.multi_layer_feed_forward(
n_input, n_output, depth = 1, width = 4))
parameter T0::Temperature = 273.15
parameter T_range::Temperature = 30
# This is a unit conversion factor
parameter C1::HeatCapacity = 1
node_a = Node()
node_b = Node()
relations
# This is some initial setup for the block
# to connect its ports.
initial node_a.T = T0
guess node_a.Q = 0.0
# The neural network operates in a specific range of numbers (0-1) for its inputs.
# You should normalize your inputs to that range. This requires that you know
# a reasonable range of values that your inputs can take.
nn.inputs[1] = (node_a.T - T0) / T_range
nn.inputs[2] = (node_b.T - T0) / T_range
# This is where we connect the neural network's output.
# Note that it is outputting heat flow here,
# and we are injecting it such that the derivative should be 0 at steady state, i.e., the component is conservative.
# Here, there is a feedback loop since one of the inputs is `node_a.T`
# and the output is related to `der(node_a.T)`.
# The neural network is learning and encoding a new state here.
C1 * der(node_a.T) = node_a.Q - nn.outputs[1]
nn.outputs[1] + node_b.Q = 0
end
component NeuralPot
parameter C2::Real = 15
heat_input = BlockComponents.Sine(amplitude=1, frequency=1/(2*pi))
source = ThermalComponents.PrescribedHeatFlow(T_ref=273.15)
pot = ThermalComponents.HeatCapacitor(C=C2, T0=273.15)
air = ThermalComponents.ThermalConductor(G=0.1)
env = ThermalComponents.FixedTemperature(T=293.15)
Tsensor = ThermalComponents.TemperatureSensor()
thermal_nn = ThermalNN()
relations
connect(heat_input.y, source.Q)
connect(pot.node, Tsensor.node)
connect(pot.node, air.node_a)
connect(air.node_b, env.node)
connect(source.node, thermal_nn.node_a)
connect(thermal_nn.node_b, pot.node)
end
analysis NeuralPotTransient
extends TransientAnalysis(stop=100.0)
model = NeuralPot()
end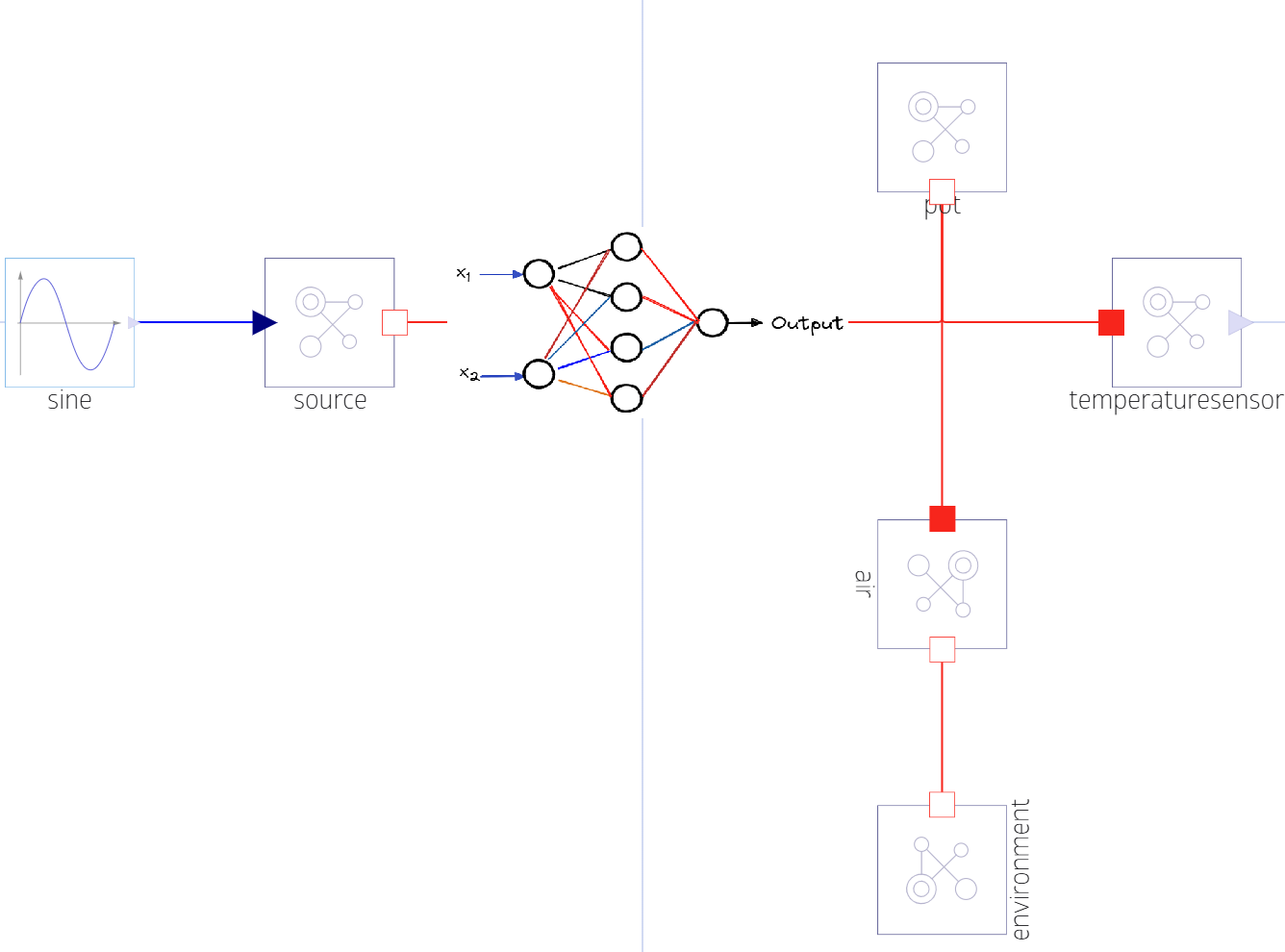
After building the ThermalNN component, we assemble the system and then define a transient analysis. Before starting the training of the neural network, we should first make sure that the augmented system is in a configuration where we can define a finite and (relatively small) loss function value.
r0 = NeuralPotTransient()
plot(r0; title = "Pre-training results")Now that we have a reasonable starting point, we can start training the network. First we will define how to train the embedded neural network using the NNTrainingAnalysis
using DyadData: DyadTimeseries
analysis NeuralPotTrainingAnalysis
extends DyadModelDiscovery.NNTrainingAnalysis(
data=DyadData.DyadTimeseries("dyad+juliahub://juliahub.com/datasets/juliasimtutorials/potplate", independent_var = "timestamp", dependent_vars = ["pot.T(t)"]),
depvars_names=["pot.T"], alg = "Tsit5", calibration_alg="SingleShooting",
optimizer="Adam", optimizer_maxiters=10000, network_component="thermal_nn.nn", results_path="r1.csv")
model = NeuralPot()
endAnd after that that, we switch to julia to do the training by running the analysis
using Main.var"##build/.dyad/analyses/udesDyadHygiene#301".component_ude # hide
r = NeuralPotTrainingAnalysis()We can now check the training results are satisfactory by inspecting artifacts such as the convergence plot
using Main.var"##build/.dyad/analyses/udesDyadHygiene#301".component_ude # hide
artifacts(r, :ConvergencePlot)and looking at the simulation results with
using Main.var"##build/.dyad/analyses/udesDyadHygiene#301".component_ude # hide
sol = artifacts(r, :CalibratedSimulation)After we are satisfied with the results, we can export them, such that we can use the neural network as a dataset for performing symbolic regression.
using Main.var"##build/.dyad/analyses/udesDyadHygiene#301".component_ude # hide
artifacts(r, :ResultsExport)We then describe the analysis in Dyad
analysis NeuralPotUDEAnalysis
extends DyadModelDiscovery.SymbolicRegressionUDEAnalysis(
data=DyadData.DyadTimeseries("dyad+juliahub://juliahub.com/datasets/juliasimtutorials/potplate", independent_var = "timestamp", dependent_vars = ["pot.T(t)"]),
depvars_names=["pot.T"], alg="Tsit5", network_component="thermal_nn.nn", training_result="r1.csv",
maxdepth = 5, maxsize=7)
model = NeuralPot()
endAnd then we go back to julia to run the analysis with
using Main.var"##build/.dyad/analyses/udesDyadHygiene#301".component_ude # hide
result = NeuralPotUDEAnalysis()Looking at the results, we can see a table of candidate models, where we can compare them based on how well they fit our data. Looking at the top candidate, we can observe that the candidate terms that would replace the neural network would be directly proportional with the difference of the neural network inputs. Going back to the definition of the ThermalNN component, we see that the inputs are the temperatures on each side of the component. Since the component outputs a heat flow rate, this also makes physical sense.
Further Reading
C. Rackauckas et al., “Universal Differential Equations for Scientific Machine Learning,” arXiv:2001.04385 [cs, math, q-bio, stat], Jan. 2020, Accessed: Feb. 09, 2020. [Online]. Available: http://arxiv.org/abs/2001.04385 ↩︎