JuliaSimSurrogates
Many industrial users unlock faster runtime when writing models in Julia, but there are cases where even this improved runtime is not enough for bottlenecks in a product's development life-cycle. When simulations are too computationally expensive or time consuming to evaluate, engineers have limited time for experimentation and exploration. In many cases, these long-running models can be replaced by much faster surrogate models, cutting down workflows from days and months to minutes and hours. This extra computational power becomes crucial for many processes like design optimization, sensitivity analysis, control tuning, etc.
However, generating surrogate models in practice requires a unique blend of knowledge in Physics, Mathematics, Scientific Machine Learning (SciML), and High Performance Computing (HPC). Traditionally, the effort and resources to generate one surrogate model for one physical system would require the skills of several modeling engineers, machine learning engineers, HPC systems engineers, and likely a couple of PhDs. After assembling this team and gaining access to the necessary compute resources, a reasonable timeline for completion is between 6 and 18 months. That's for one surrogate.
Why JuliaSimSurrogates?
With JuliaSimSurrogates, a single person or a team is empowered to carry out all that work in far less time, and without that level of expertise. Other benefits include:
Theoretical Soundness: Designed by experts in the field,
JuliaSimSurrogatesgoes beyond the state of the art in SciML methods and itself sets the bar for what is truly possible in the domain. It incorporates all the right inductive biases that are generic enough to accommodate all domains of simulation, while generalising well with theoretical guarantees in terms of accuracy and performance as the number of data points used to train the surrogate increases.Tight Integration Industry Standard Modelling Tools: Integrate fast surrogates back into your simulation environment of choice as a drop-in replacement using FMUs. Further, integration with ModelingToolkit.jl and SciML gives access to bespoke high-performance solvers for all types of problems that aren't found anywhere else - JuliaSimSurrogates seamlessy leverages the latest and greatest in model building and simulation as well as its continued fast-paced development.
Speed that cannot be found anywhere else: With Julia as the programming language that JuliaSimSurrogates is built with, performance gains of fast code with the speedup a surrogate brings an unmatched level of performance that will enable workflows never before possible.
Examples
Saying something is 25 to 1000x faster that is just as accurate as your realistic model can be difficult to comprehend or visualize. In the spirit of helping one visualize and understand these performance gains, a few examples that demonstrate how surrogates can be generated and deployed to solve computationally expensive problems at scale using JuliaSimSurrogates can be found below. This list is constantly growing and is only the beginning.
1. Brusselator
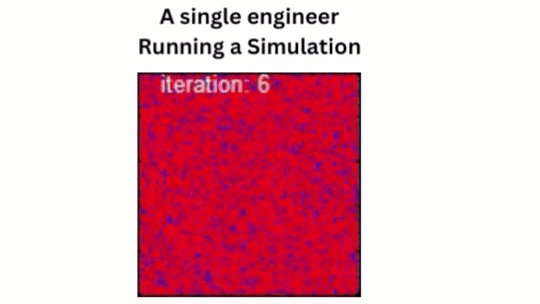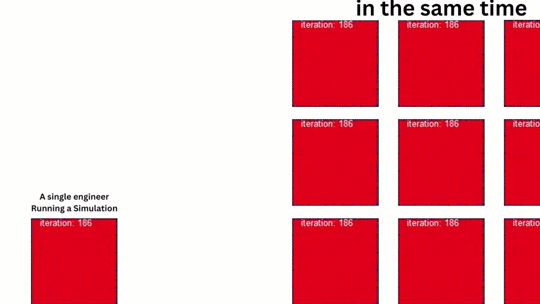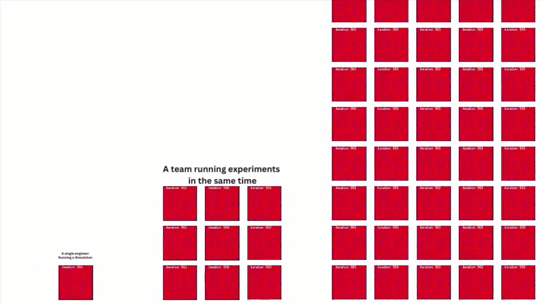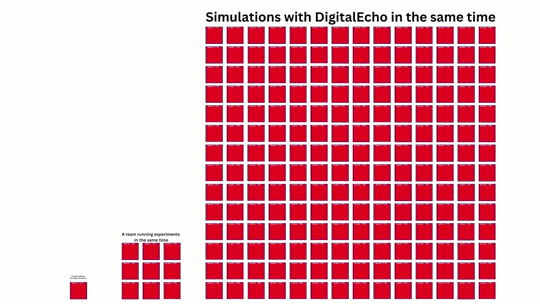
The Brusselator model is a stiff PDE that is used to model the dynamics of a chemical reaction system. In practice, fine discretizations of this model result in solve times that can take minutes to hours. Given someone wanted to perform some kind of parmaeter estimation using it, 1000's or 10000's of solves would be required, leading to waiting periods that could easily turn into weeks. This difficulty is further compounded by the fact that using machine learning to learn a stiff model is incredibly difficult and not solved at scale in the current open literature. In this showcase, it will be demonstrated how a single person using JuliaSimSurrogates is empowered to not only generate a surrogate for a stiff system, but one that is incredibly fast, leading to an output that teams of people using the original model could not rival.
2. Battery
With electrification on the rise, simulating batteries and their ability to meet the demands of the loads they serve in the real world can be very computationally expensive. As batteries grow in size, the simulation speed scales very poorly despite the dynamics not varying much. This prevents workflows that use real world voltage demands from electric vehicles to simulate them for larger battery packs viable in practice. Furthermore, how to generate data for these kinds of systems so that a surrogate can be trained and reliably used without many examples of the kind of inputs to expect is non-trivial and unsolved at scale as well. In this showcase, it will be demonstrated how a single person is empowered with the ability to not only generate a battery surrogate that can act as a reliable, high performance stand in for the original slow battery model with unseen voltage inputs, but also show how easy it is made for the user through JuliaSimSurrogates to generate data for it.
Getting Started
With JuliaSimSurrogates.jl, all a user has to do is:
- Configure your data generation process using
SimulatorConfig. This will run your simulator through all the different situations you configured on the cloud, and in a fully parallelized manner.
Wrap a
DigitalEchoto the results of theSimulatorConfigonce it is done running all the simulations. This isJuliaSimSurrogates'automated surrogate generation technology.Validate results through visualizations designed to give full visibility of the surrogates performance.
Pass surrogate to JuliaSimSurrogates Deployment API which will wrap it into an FMU in an automated manner
With this kind of workflow, the user is expected to only configure the kind of behaviour the simulator should generate data for, and JuliaSimSurrogates will create a DigitalEcho of it! Preprocessing, model specification and training are all done on behalf of the user leaving only the duty of checking if the surrogate meets the performance standard for the intended use case.
To learn more about the open source ecosystem, please checkout the official Julia Lang documentation and the SciML Open Source Software Ecosystem documentation.
JuliaSim is a proprietary software developed by JuliaHub Inc. Using the packages through the registry requires a valid JuliaSim license. It is free to use for non-commercial academic teaching and research purposes. For commercial users, license fees apply. Please refer to the End User License Agreement for details. Please contact sales@juliahub.com for purchasing information.